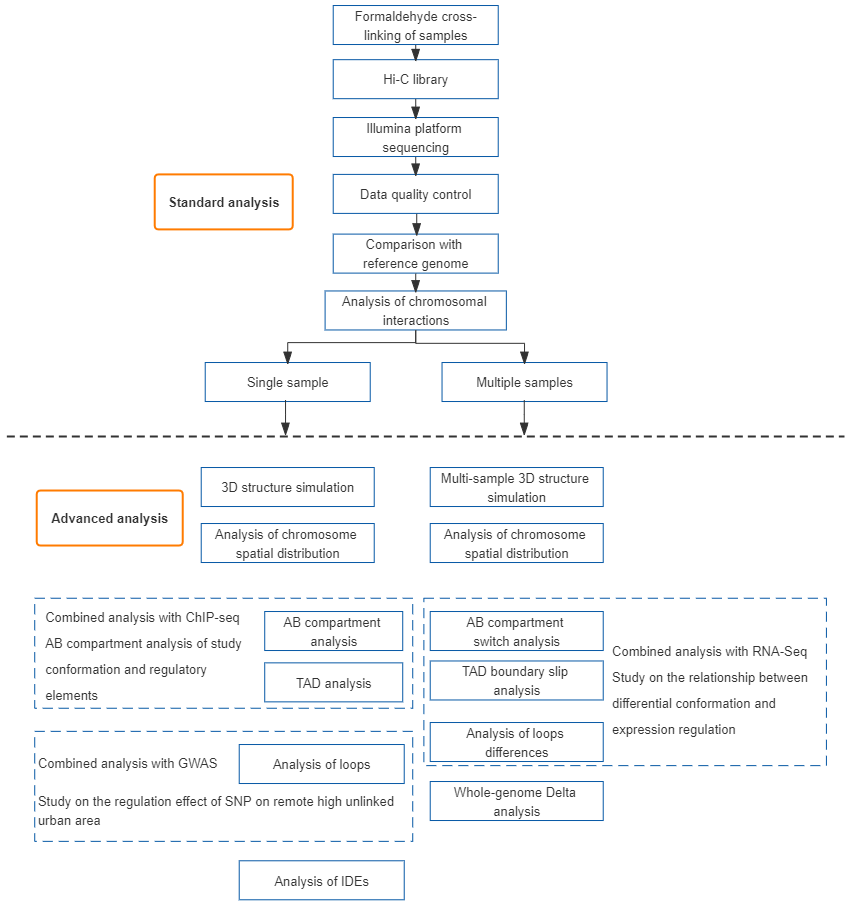
Hi-C technology is derived from the technology of Chromosome Conformation Capture-3C, takes the entire nucleus as the research object, adopts high-throughput sequencing technology, and combines bioinformatics methods to study the spatial relationship of the entire chromatin DNA in the whole genome, obtain high-resolution three-dimensional structure information of chromatin by capturing all DNA interaction patterns inside, and analyze it with ATAC-seq, ChIP-seq, genome, transcriptome data, etc., thereby elucidating the related mechanism of organism trait formation from gene regulatory network and epigenetic network.